Improving Binder Success Rates with BindCraft
We’re excited to announce that BindCraft1 has been added to our toolset at Levitate Bio! This protocol has huge potential for improving de novo binder design by enhancing success rates, stability, and affinity for target proteins.
RFDiffusion has successfully generated many de novo binders, but its hit rate for binding could use improvement and it often requires additional design rounds to achieve optimal affinity. An alternative approach is to start with a native binding site and use RFDiffusion to build a novel protein around the binding protein. However, the latest AI tool from the Rosetta community is changing the game—producing strong binders with a high success rate, even without relying on a native binding protein.
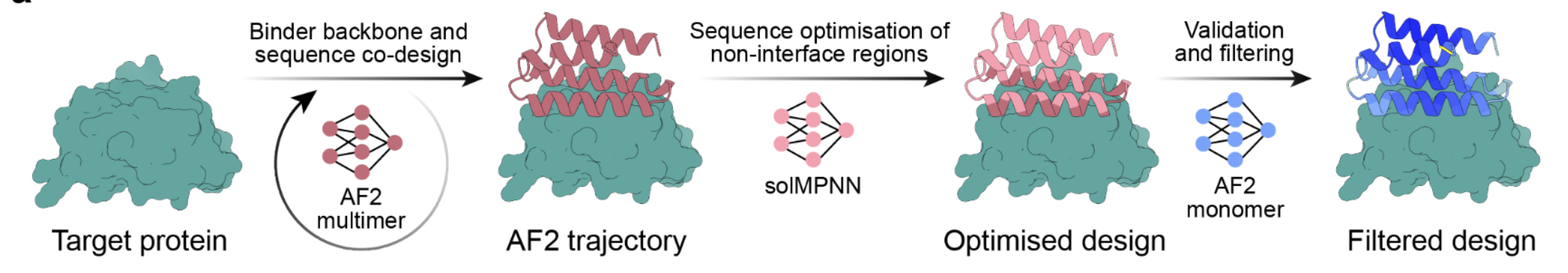
Overview of the BindCraft Protocol
BindCraft modifies the ColabFold implementation of AlphaFold2 Multimer (AF2M) to generate novel binding proteins for completely new binders. It also uses ProteinMPNN and PyRosetta to improve and filter the AF2M binders. BindCraft enables the design of fully de novo proteins that are stable, soluble, and highly effective at binding their targets.
How BindCraft Works
- Define Input Parameters
- Provide target protein structure & sequence.
- Set binder length range & secondary structure bias (e.g., helices vs. β-sheets).
- Optionally specify a desired binding site.
- BindCraft generate a Random Sequence & Initial Structure Prediction with the target
- BindCraft starts with a completely random sequence for the binder.
- AlphaFold2 Multimer (AF2M) co-folds the binder with the target.
- This is expected to produce a very poor initial structure but give BindCraft a starting point that it will try to improve.
- Iterative Sequence Optimization & AI-Guided Refinement
- The initial binding model is scored using multiple metrics, including:
- AlphaFold2 confidence scores
- Interface interaction strength
- Helical/beta-sheet balance
- Binding site alignment (if provided)
- Backpropagation & AI refinement suggest new sequences to improve binding.
- Backpropogation runs AF2M in reverse to predict what sequence variation would make the scores better.
- BindCraft uses gradient-based optimization and sequence backpropagation to iteratively refine binder sequences.
- Repeat the prediction-scoring-refinement cycle until:
- A strong binder emerges, OR
- The model aborts unsuccessful designs.
- The initial binding model is scored using multiple metrics, including:
- Final Design Refinement & Filtering
- The binder created by AF2M must be further optimized because AF2 performs poorly at creating soluble proteins.
- BindCraft uses ProteinMPNN to redesign the binder, excluding the binding site residues.
- New sequences are folded as monomer to validate that sequence forms predicted fold with target protein for validation and filtering
- Filters out designs that:
- Score below AF2M confidence threshold (~0.7-0.8).
- Have <7 interface residues or structural clashes.
- Top candidates are moved to wet-lab testing.
When is BindCraft a Better Choice Than RFdiffusion?
Both BindCraft and RFdiffusion are powerful tools for binder design, but BindCraft can be more effective in certain cases due to key advantages:
- Cofolding for Better Optimization
- BindCraft cofolds the binder and target together, optimizing both structure and sequence simultaneously.
- RFdiffusion generates a rigid binder first, then ProteinMPNN fits a sequence, which may lead to suboptimal interactions.
- Best for: Flexible targets, induced fit interactions, intrinsically disordered regions (IDRs).
- Secondary Structure Control
- BindCraft allows biasing toward different secondary structures, overcoming RFdiffusion’s alpha-helix bias.
- Best for: Designing binders with diverse folds, such as beta-sheet-rich interfaces.
- Avoids Unrealistic Structures
- RFDiffusion’s stochastic diffusion process can create poorly packed or unphysical designs.
- BindCraft integrates structure-based refinement, reducing hallucinations.
- Best for: Generating experimentally viable, well-packed binders.
- Better Sequence-Structure Compatibility
- BindCraft optimizes sequence and structure together, leading to stronger binding affinities.
- RFdiffusion is used to generate structures predicted to bind a target, but much of its output will not be good binders and many steps are needed to identify the best hits.
- Best for: Designing binders with precise binding interactions.
Experimental Results: How Well Does BindCraft Work?
BindCraft was tested on 10 different target proteins. For each, dozens of binders were experimentally tested via SPR (Surface Plasmon Resonance). The image below highlights the # of Binders found / # tested for each target and reports the best Kd of the binders for each target.
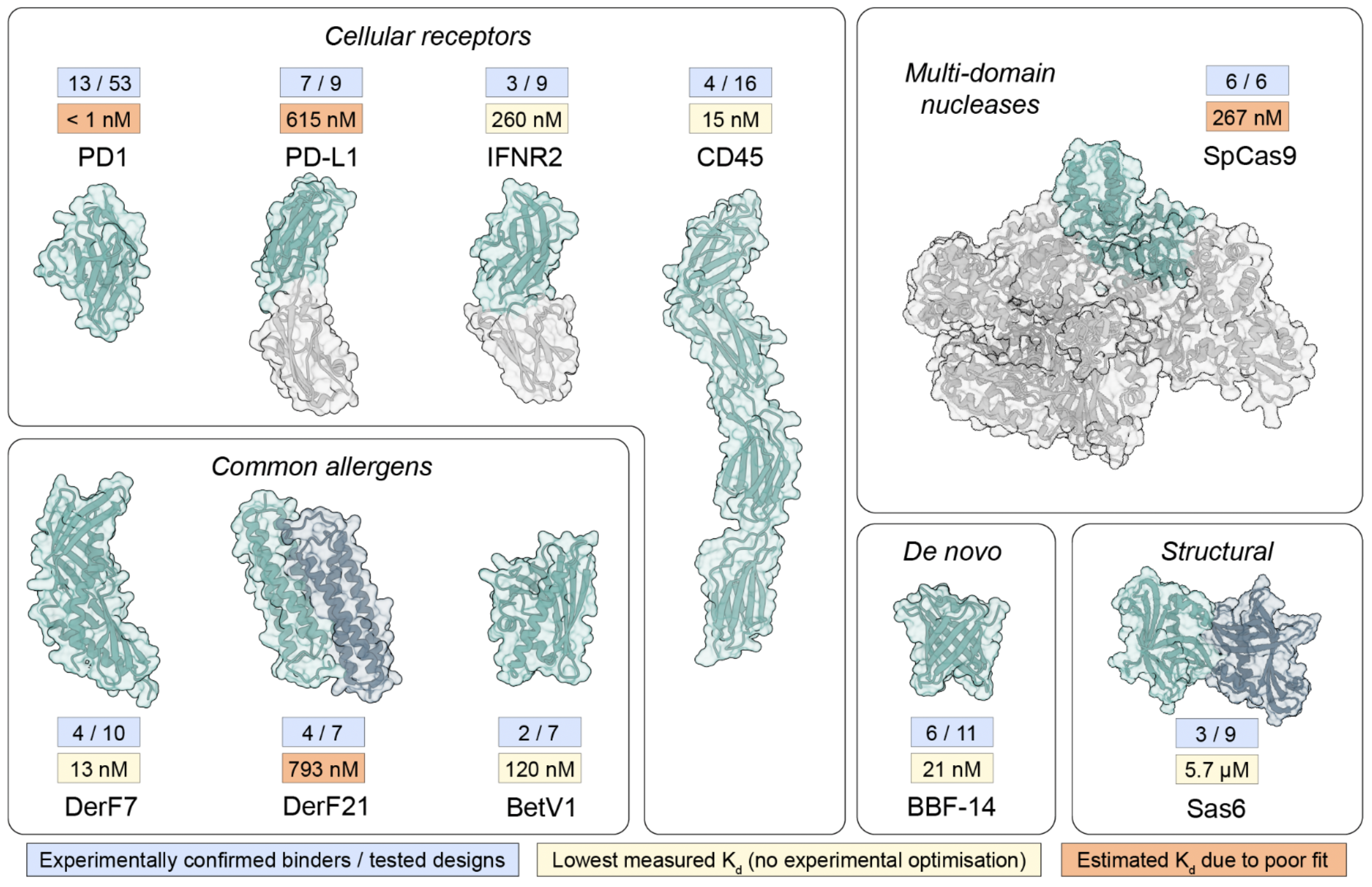 Pacesa et al.1 Figure 1
Pacesa et al.1 Figure 1
Why This Is Impressive
- These hit rates are significantly better than most de novo binder design methods.
- The binding affinities (Kd values) show that many binders achieved sub-nanomolar affinity, indicating strong binding.
Who Developed BindCraft?
BindCraft was developed through a collaboration between two Rosetta labs; the Correia Lab (EPFL, Switzerland) and the Ovchinnikov Lab (MIT).
This partnership combined deep expertise in AlphaFold-based structure prediction and computational protein design, resulting in a new, highly effective pipeline for de novo binder generation.
Why this Matters & What’s Next
BindCraft is a major advance in de novo protein design. It enables:
✅ Binder creation from scratch, eliminating the need for pre-existing motifs or extensive optimization.
✅ Automated sequence refinement, significantly improving success rates.
✅ Better experimental success rates, making de novo design more cost-effective.
At Levitate Bio, we have already integrated BindCraft into our computational binder design pipeline. We are excited to apply this approach to challenging protein targets where traditional de novo design struggles.
If you’re working on a protein-targeting problem and want to explore BindCraft-driven designs, reach out to us!
References
-
Martin Pacesa, Lennart Nickel, Joseph Schmidt, et al. BindCraft: one-shot design of functional protein binders. https://doi.org/10.1101/2024.09.30.615802 ↩ ↩2