Mitigating Manufacturing Liabilities with Post-Translational Modification Prediction
Maintaining therapeutic stability and potency during manufacturing and storage continues to be a costly and significant challenge in development. Protein therapeutics are susceptible to multiple forms of degradation and can host significant liabilities as a result of post-translational modification (PTM).
Degradation may result from asparagine (Asn) deamidation, aspartate (Asp) isomerization, and methionine (Met) oxidation, as well as significant risk associated with aggregation and viscosity during administration and manufacturing1. Early screening for liabilities of potential therapeutic candidates is critical in reducing development costs and ultimately enabling downstream success, either by allowing for early manufacturing and formulation adjustments or remedying problematic features in the protein2. Thus, in silico efforts for predicting chemical liabilities during early design and discovery hold high interest in reducing costs and improving efficacy1.
Currently, companies such as Schrödinger and Chemical Computing Group (CCG) offer tool suites that provide metrics to account for potential liabilities in protein design. Common industry assessments rely solely on sequence-based PTM motif scans which often over-predict liabilities and lack any structural data necessary for better predictions.
In contrast, Levitate Bio’s ptm-prediction tool implements state of the art machine learning models and standard industry methods for predicting susceptible residues for PTMs of concern, where predictions are informed by a combination of both structural and sequential features extracted from provided inputs.
By combining conventional sequence-based analysis with structural features in optimized machine learning models, Levitate Bio is able to predict most PTMs of concern, achieving high performance in recall and prediction overall.
Within minutes, Levitate Bio’s ptm-prediction API provides users with easy and ready to use results for reporting on potential chemical liabilities. Along with a formalized report of detected PTMs of concern and potential mitigation strategies, the API will also provide an easy to use PyMOL script for visualizing detected PTMs on a provided structure. Offering both speed and performance, the ptm-prediction API provides essential insights on possible liabilities for proteins at any stage in development.
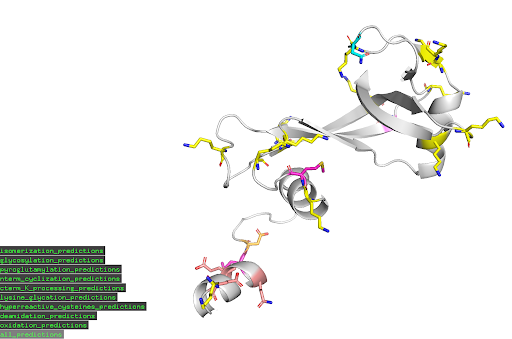
Example PyMOL visualization of identified PTMs of concern for provided input PDB structures
PTMs Covered by Levitate Bio’s ptm-prediction
- Asparagine Deamidation
- Aspartic Acid Isomerization
- Methionine Oxidation
- Hyper-reactive Cysteines
- N-Linked Glycosylation
- Lysine Glycation
- Pyroglutamylation
- N-Term Cyclization
- C-Term Lysine Processing
References
-
Sankar, K., Hoi, K. H., Yin, Y., Ramachandran, P., Andersen, N., Hilderbrand, A., McDonald, P., Spiess, C., & Zhang, Q. (2018). Prediction of methionine oxidation risk in monoclonal antibodies using a machine learning method. MAbs, 10(8), 1281–1290. https://doi.org/10.1080/19420862.2018.1518887 ↩ ↩2
-
Yang, X., Xu, W., Dukleska, S., Benchaar, S., Mengisen, S., Antochshuk, V., Cheung, J., Mann, L., Babadjanova, Z., Rowand, J., Gunawan, R., McCampbell, A., Beaumont, M., Meininger, D., Richardson, D., & Ambrogelly, A. (2013). Developability studies before initiation of process development. MAbs, 5(5), 787–794. https://doi.org/10.4161/mabs.25269 ↩