ThermoMPNN API
The ThermoMPNN API provides an interface to the AI based ThermoMPNN protocol for performing a full site saturation mutagenesis (SSM) DDG analysis of a protein. A full SSM is generated even if you are only interested in 1 mutation as the entire protocol runs on the order of seconds vs the 10s of minutes per mutation of the older Cartesian DDG tool.
Command Line Interface
Examples
Perform a full SSM
lev engine submit thermo-mpnn input.pdb
Perform a full SSM on a specific chain
lev engine submit thermo-mpnn input.pdb --chain A
Perform a full SSM on a specific chain using the modified alphabet
lev engine submit thermo-mpnn input.pdb --chain A --alphabet QENHDRK
Flags
--alphabet(bool) (Optional)- Whether to use the standard amino acid alphabet or the modified alphabet used in the original ThermoMPNN paper. ng the structure you would like to perform the SSM on.
--chain(str) (Optional)- The chain ID of the structure to perform the SSM on.
--gpu-type(str) (Default:t4)- The type of GPU to use for the job.
- Options:
t4a100
--pdb-file(str) (Required)- The pdb file containing the structure you would like to perform the SSM on.
Python Interface
Examples
Perform a full SSM:
from engine import EngineClient
client = EngineClient()
client.authorize()
job_id = client.submit_thermo_mpnn(
pdb_file="input.pdb"
)
Perform a full SSM on a specific chain:
job_id = client.submit_thermo_mpnn(
pdb_file="input.pdb",
chain="A"
)
Flags
pdb_path(str) (Required)- The pdb file containing the structure you would like to perform the SSM on.
chain(str) (Optional)- The chain ID of the structure to perform the SSM on.
gpu_type(str) (Default:t4)- The type of GPU to use for the job.
- Options:
t4a100
Outputs
predicted_ddgs.csv(csv file)- A csv file containing the DDG predictions for every mutation.
In-depth
Introduction
ThermoMPNN is a deep learning tool for in silico prediction of protein stability changes due to single point mutations. With accuracy rivaling or surpassing other tools, ThermoMPNN stands light years ahead in speed. Test every conceivable point mutation in large proteins within minutes. ThermoMPNN, an evolution of ProteinMPNN, takes protein sequence generation to the next level. Initially trained on 19,700 protein clusters from the PDB, ProteinMPNN excelled in generating sequences with high native recovery rates. However, as stability is multifaceted, ThermoMPNN integrates stability data for enhanced bias for protein thermostability, redefining the tool’s training approach to prioritize stability in sequence generation.
For ThermoMPNN, they transfer some of the learning from ProteinMPNN to use as input for ThermoMPNN. They utilized its sequence recovery and trained the model with a large set of experimental stability measurements to teach ThermoMPNN to predict the stability of single point mutations on a wild-type structure. The experimental data came from a groundbreaking study that took 1.8 million measurements in total to calculate thermodynamic folding stability for over 300 proteins with every point mutation. This “Megascale dataset” helped ThermoMPNN learn generalizable structural determinants of stability.
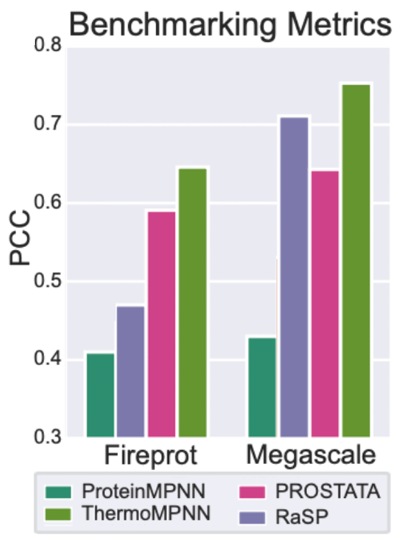
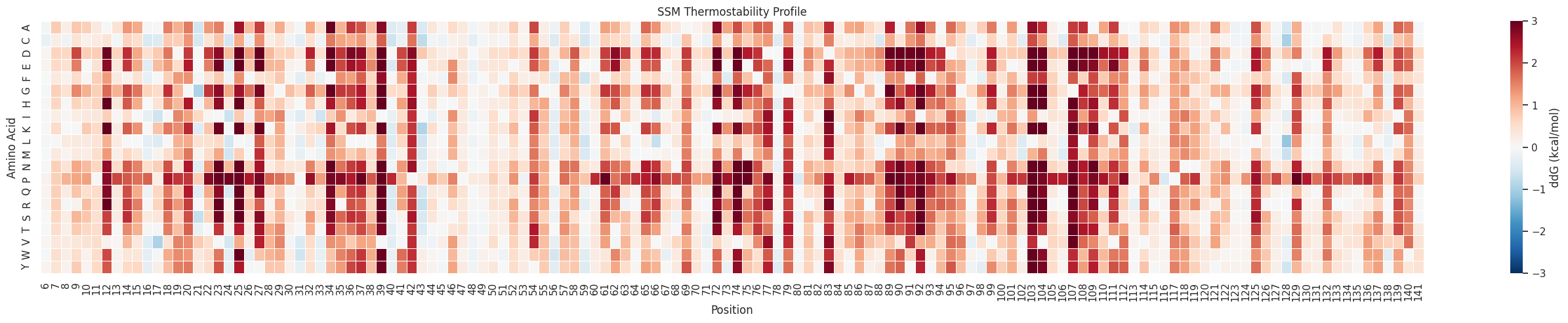
ThermoMPNN outputs the predicted ΔΔG° (kcal/mol) for mutation to each possible amino acid at every possible position. ThermoMPNN was benchmarked against 3 other tools using the FireProt data set as well as the Megascale set. It was statistically superior for both data sets in terms of Comparison of Spearman (SCC, right top) and Pearson (PCC, right bottom) correlation coefficients. The Megascale set came from the same experiment as the training data, but the test set did not overlap and homologs were removed.
Although it represents an improvement, the developers have identified certain trends indicating biases toward specific amino acids. Notably, there is a preference for hydrophobic mutations, even at surface positions. Particularly, there is an elevated affinity for isoleucine, tryptophan, and surface arginine, coupled with a decreased preference for surface lysine and glutamic acid residues. The tool exhibits slightly greater accuracy for proteins under 100 residues and demonstrates superior performance on native proteins compared to designed proteins which is true for more in silico tools.
How to run
Running ThermoMPNN only has a single command.
lev engine submit thermo-mpnn example.pdb
This will produce a csv file with a full site saturation mutagenesis (every position mutated to every amino acid) along with the predicted change in free energy associated with that mutation.
References
- Robust deep learning-based protein sequence design using ProteinMPNN. Dauparas J, Anishchenko I, Bennett N, Bai H, Ragotte RJ, Milles LF, Wicky BIM, Courbet A, de Haas RJ, Bethel N, Leung PJY, Huddy TF, Pellock S, Tischer D, Chan F, Koepnick B, Nguyen H, Kang A, Sankaran B, Bera AK, King NP, Baker D. Science. 2022 Oct 7;378(6615):49- 56.
- Transfer learning to leverage larger datasets for improved prediction of protein stability changes. Dieckhaus H, Brocidiacono M, Randolph N, Kuhlman B. BioRxiv. 2023 Jul 30:2023.