ProteinMPNN
OVERVIEW
ProteinMPNN is a sequence design tool that rapidly generates new sequences predicted to fold to the backbone of the input structure. ProteinMPNN utilizes a deep-learning neural network to interpret the backbone coordinates of a protein structure and design novel sequences.
When recombinantly expressed, these sequences often have incredible solubility and stability, making ProteinMPNN a great tool for applications in synthetic biology and therapeutic optimization.
Our implementation of ProteinMPNN in Bench makes both setup and interpretation of the resulting sequences fast and easy.
To learn more about advanced uses of ProteinMPNN, the GitHub repository provides many in-depth examples.
RUNNING ProteinMPNN
-
To begin, open an existing project or create one in Bench.
-
Click on a structure collection on the left in the Structures panel, or load one using the Structure Loader button. Once you have selected the desired structure to run proteinMPNN on click the proteinMPNN button in the Actions panel to open a jon setup tab.
-
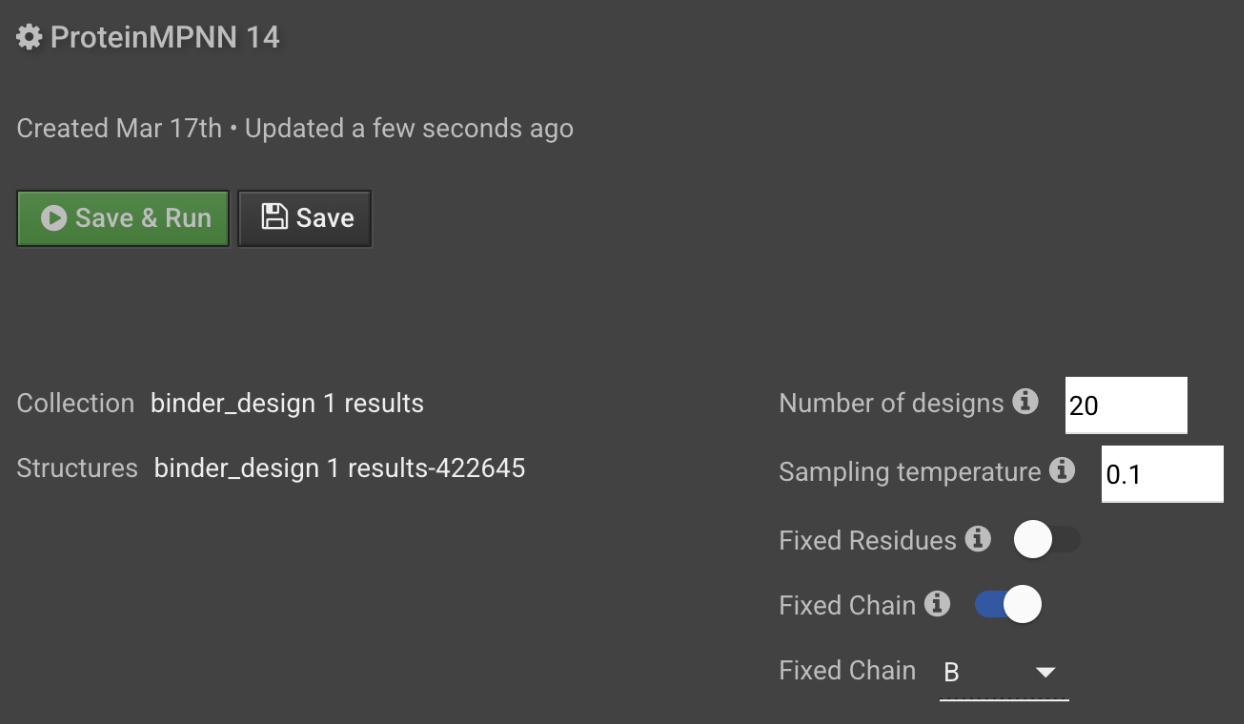
Enter the number of designs you want to generate. Protein design protocol typically generate 10-20 sequences per structure.
-
Optional: Modify the sampling temperature. Increasing the sampling temperature increases the amount of noise incorporated during sequence generation, which in turn increases the diversity of resulting sequences.
-
If you want to retain function or activity but increase stability and solubility, you can fix certain residues or chains necessary for function or activity.
- Toggle Fixed Residues ON and choose the Selector of the positions in your input structure that you do not want to change.
OR
- Toggle Fixed Chain ON and select the chain that you do not want to change. During de novo binder design, for instance, you would select the target chain, allowing only the binder to be designed by ProteinMPNN.
- Select Save & Run to start the job.
Once the job completes you will see an output Sequence collection created. Sequences within this collection can be selected, viewed as sequence logos, and used as input to run AI Folding jobs.
References
Robust deep learning–based protein sequence design using ProteinMPNN